Part:BBa_K2371009
sgRNA generator for EML4-ALK variant B 126
The echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) is a fusion gene resulting from chromosome inversion and was identified in nonsmall cell lung cancer(NSCLC).The EML4-ALK fused gene has two variants (A and B).Using computer stimulation, we selected seven sgRNA binding sites from all the combination of possible sites. Four sgRNA binding sites on Variant A and three sgRNA binding sites on Variant B.
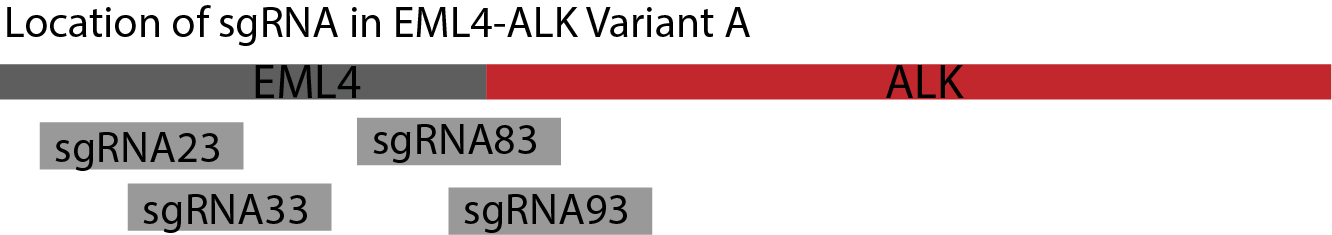
Figure 1. The four sgRNA binding sites of fusion gene EML4-ALK Variant A. We designed four sgRNA binding sites in target gene – sgRNA23, sgRNA33, sgRNA83 and sgRNA93. The number of them indicates the location of their first base pair in target gene. The combination of sgRNA binding sites will be based on the rule that a pair of sgRNA should be distributed in EML4 or ALK area separately or one of them located on the connecting area of the EML4-ALK fusion gene.
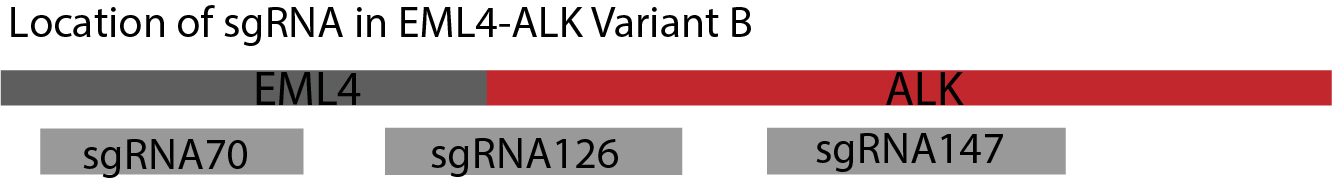
Figure 2. The three sgRNA binding sites of fusion gene EML4-ALK Variant B. We designed three sgRNA binding sites in target gene – sgRNA70, sgRNA126 and sgRNA147. The number of them indicates the location of their first base pair in target gene. The combination of sgRNA binding sites will be based on the rule that a pair of sgRNA should be distributed in EML4 or ALK area separately or one of them located on the connecting area of the EML4-ALK fusion gene.
This specific sgRNA generator is to produce sgRNA in vitro for detection of EML4-ALK cancer gene in pair with BGIC-Union NT7-dCas9(BBa_K2371000) and CT7-dCas9(BBa_K237101).
This part is designed with T7 promotor and T7 terminator and can effectively generate specific RNA in vitro. The product can have RNA concentration of 1500 ng/ul or higher with MEGAscript™ T7 Transcription Kit and after the purification and collection using Ambion® mirVana™ miRNA Isolation Kit.

Figure 3. The plasmid of sgRNA generator. The sgRNA generator Biobrick is composed of a T7 promotor, coding sequence for sgRNA and a T7 terminator.
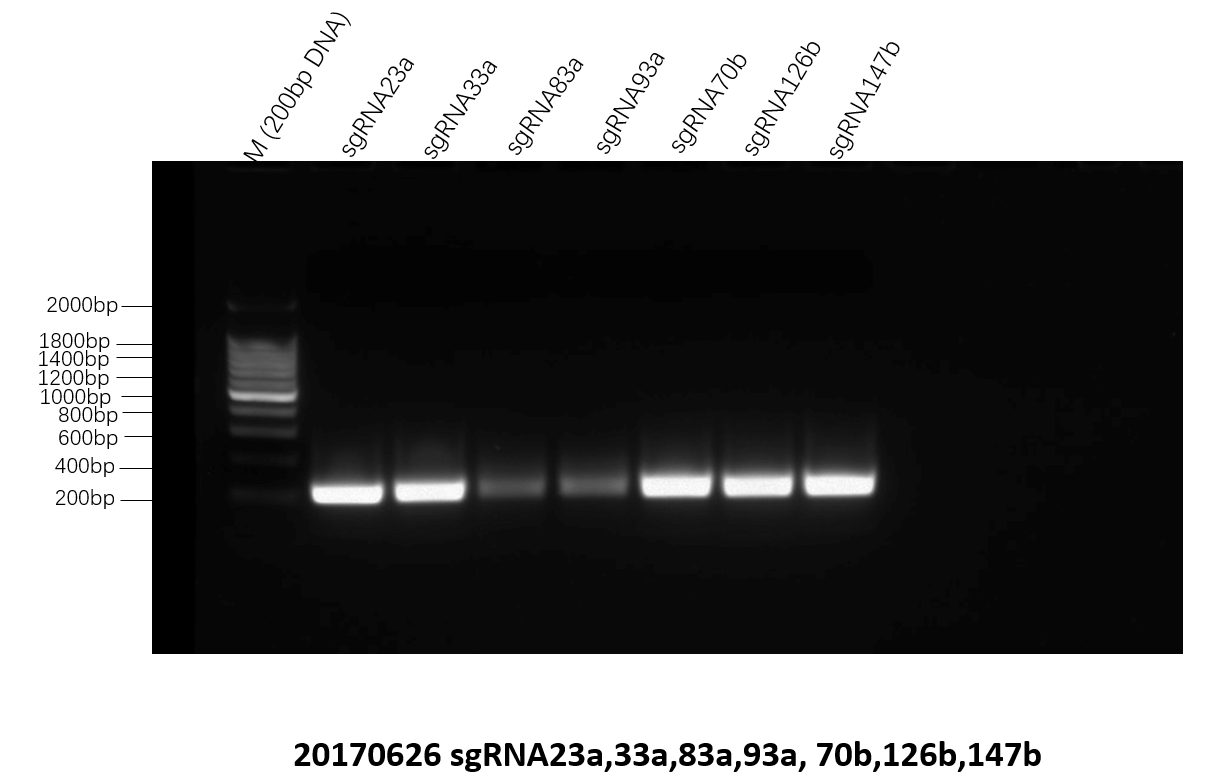
The sequence of enzyme sites, T7 promotor, sgRNA sequence and T7 terminator were synthesized by SOEPCR designed by DNAworks. The gel electrophoresis of the second amplification in SOEPCR using the oligo DNA in the start and the end of the sequence as primers validate the success of the synthesis.
Figure 4. The electrophoresis of second amplification using the using the oligo DNA in the start and the end of the sequence as primers.The desired length should be around 200bp.
Then we inserted each of those seven parts into psb1c3 plasmid.
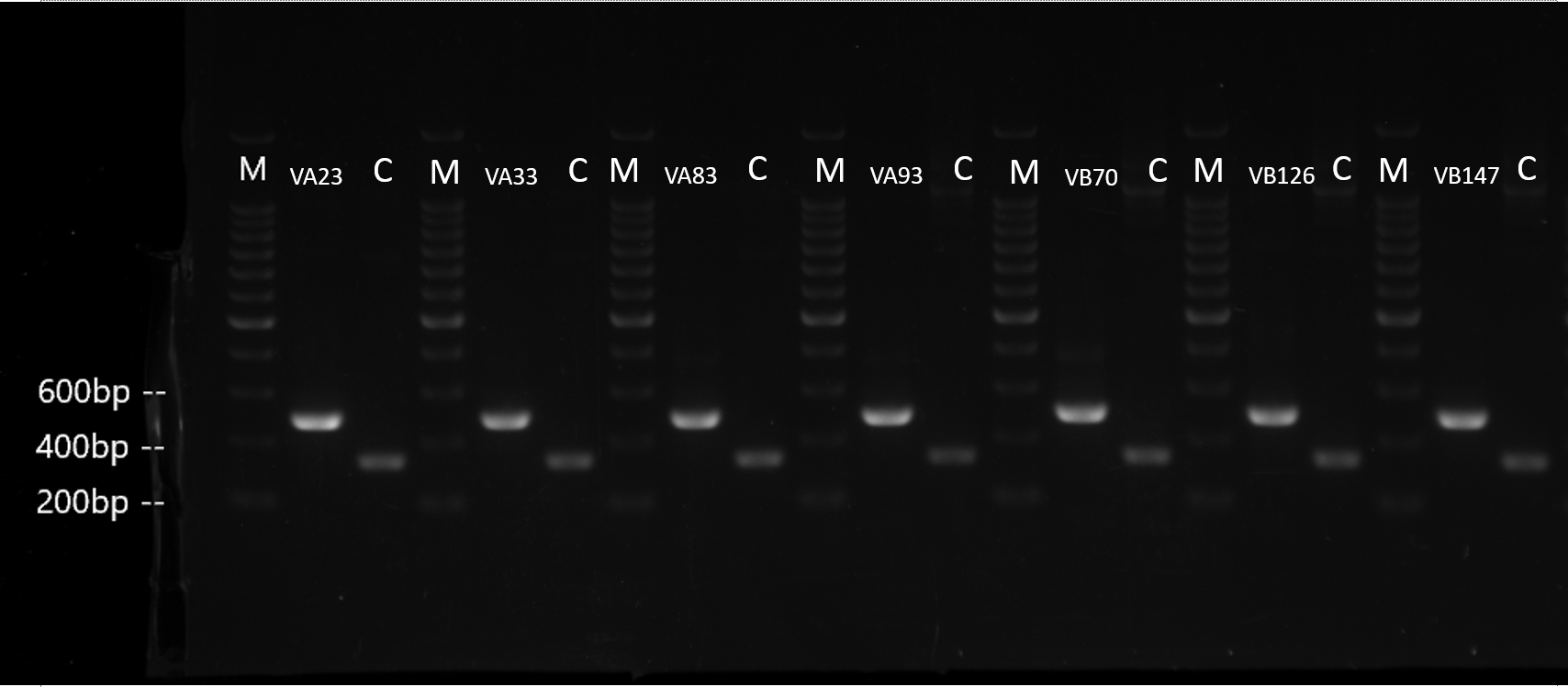
Enzyme check(XbaI and SpeI) and PCR(using VF and VR) check were conducted to prove the success of connection.
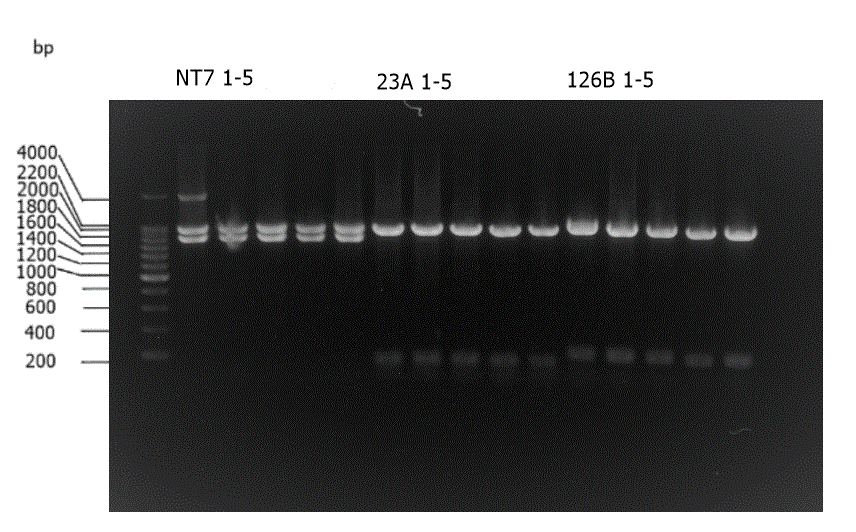
Figure 5. Enzyme check of sgRNA generator variant B 126. XbaI and SpeI were used. The expected length should be around 200bp.
Figure 6. PCR check of seven sgRNA generators. The PCR check is conducting using VF and VR as primers. Marker is noted as M which has a ladder of 200bp. Control is a J23119 biobrick and noted as C. PCR result of control should be about 350bp. VA23 stands for sgRNA generator for EML4-ALK Variant A 23 and so on. The length of the PCR result of sgRNA generator should be about 500bp.
In order to synthesis RNA, we conducted a PCR to linearize the sequence. Then the purified PCR products were transcribed and purified in vitro.(Protocol ← click here)
Finally a RNA electrophoresis was conducted to corroborate the existence of RNA.
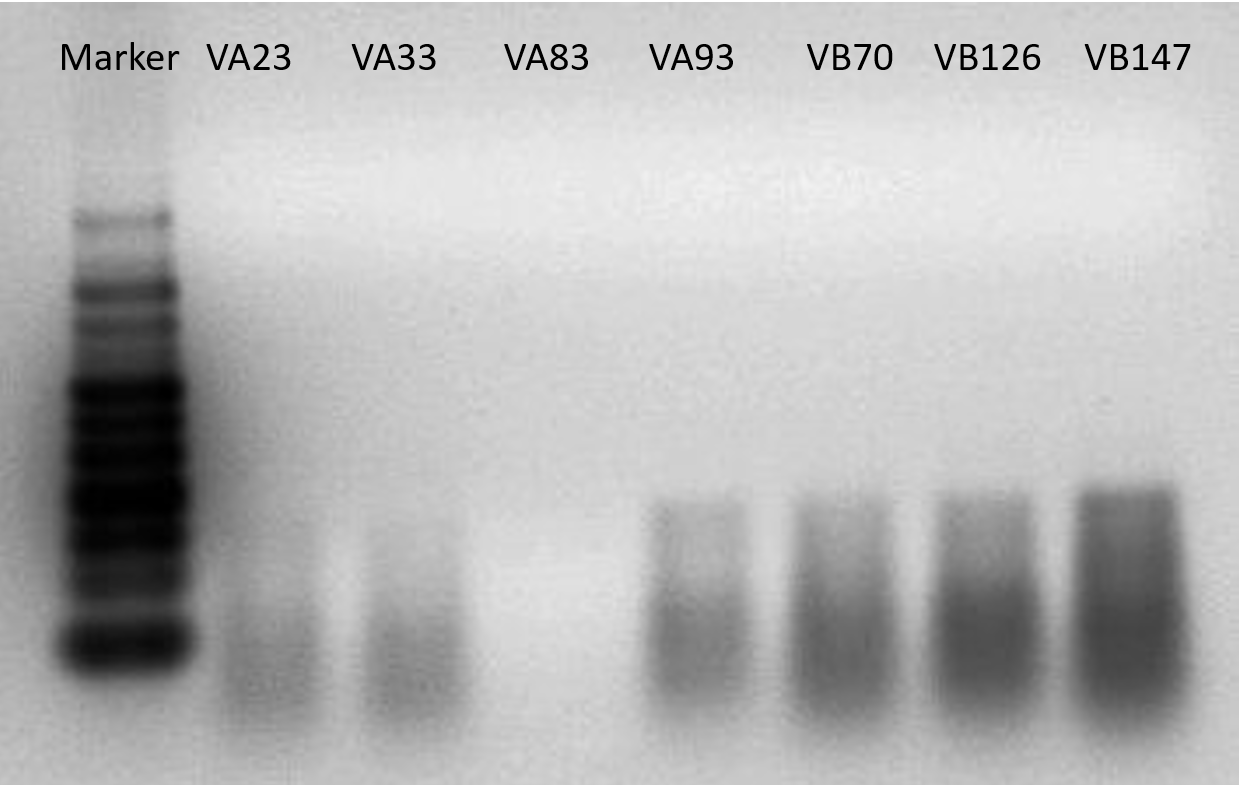
Figure 7. Native RNA electrophoresis of all the seven sgRNA. Because our RNA electrophoresis is native electrophoresis, the each RNA won’t concentrate to a band since they may form higher structure. The only purpose we did it was to ensure we had successfully obtain the sgRNA.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |